A Minimum Demo
Here’s a minimum demo to get started with ELLA.
Install ELLA
Install ELLA follows the steps in Install ELLA if you haven’t done so yet.
The script and data that will be used in this demo should have already been downloaded (while cloning the ELLA repo). You should be able to find these at your local ELLA folder:
ELLA/scripts/demo/mini_demo/
├── lightning_logs
│ └── run1
├── log
├── mini_demo_data.pkl
├── prepared_data
│ ├── cells_center.json
│ ├── cells_point_infos.json
│ ├── cells_polygon.json
│ └── training_data.jsonl
└── run_mini_demo.sh
The input data (mini_demo_data.pkl) mainly contains a dictionary of three dataframes corresponding to gene expression, cell segmentation, and nucleus segmentation (optional). The data contains 5 cells and 4 genes, and its details are explained in ELLA’s Inputs.
ELLA Anlysis
1. We first process the input data. This step includes registering cells (computing relative positions) and restructuring the data for efficient cell-type and gene-level parallelization.
python -m ella.data.prepare_data -i your_dir/ELLA/scripts/demo/mini_demo/mini_demo_data.pkl -o your_dir/ELLA/scripts/demo/mini_demo/prepared_data
The outputs including cells_center.json, cells_point_infos.json, cells_polygon.json, and training_data.jsonl.
2a. We can run one gene on a local machine by training a ELLA model based a recipe e.g. ELLA/configs/mini_demo.yaml
ella-train --config-name mini_demo
2b. We can run multiple genes in parallel using scripts e.g. run_mini_demo.sh on a computing cluster.
bash run_mini_demo.sh
Check out ELLA’s results
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
red = '#c0362c'
lightorange = '#fabc2e'
lightgreen = '#93c572'
lightblue = '#5d8aa8'
darkgray ='#545454'
colors = [red, lightorange, lightgreen, lightblue]
nr = 1
nc = 4
ss_nr = 1.7
ss_nc = 2
fig = plt.figure(figsize=(nc*ss_nc, nr*ss_nr), dpi=300)
gs = fig.add_gridspec(nr, nc,
width_ratios=[1]*nc,
height_ratios=[1]*nr)
gs.update(wspace=0.3, hspace=0.5)
res_dir = 'your_dir/ELLA/scripts/demo/mini_demo/lightning_logs/run1'
p_cauchy = []
for i in range(4):
path = f'{res_dir}/gene_{i}_estimation_result.json'
if os.path.exists(path):
with open(path, "r", encoding="utf-8") as f:
res = json.load(f)
gene_id = res['gene_id']
lam_est = res['lam_weighted']
p_cauchy.append(res['p_cauchy'])
ax = plt.subplot(gs[0,i])
ax.set_title(gene_id)
lam_std = (lam_est-np.min(lam_est))/(np.max(lam_est)-np.min(lam_est))
ax.plot(np.linspace(0,1,len(lam_std)), lam_std, lw=2, color=colors[i])
ax.set_xticks([0,0.5,1], [0,0.5,1])
ax.set_yticks([0,0.5,1], [0,0.5,1])
ax.set_xlabel('Relative Position')
if i==0:
ax.set_ylabel('Expression Intensity')
Compute FDR-corrected P values
reject, p_fdr, _, _ = multipletests(p_cauchy, alpha=0.05, method='fdr_by')
print(np.sum(p_fdr<=0.05))
print(p_fdr)
Prints:
4
[2.31296463e-16 0.00000000e+00 4.62592927e-16 2.38025062e-04]
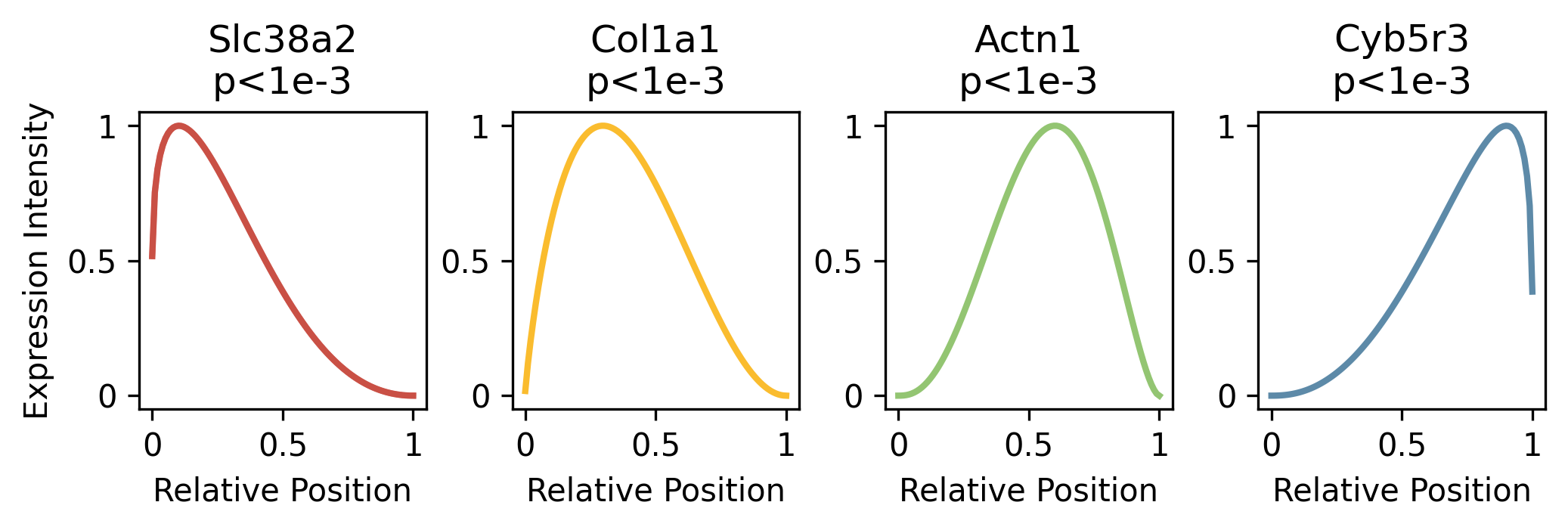
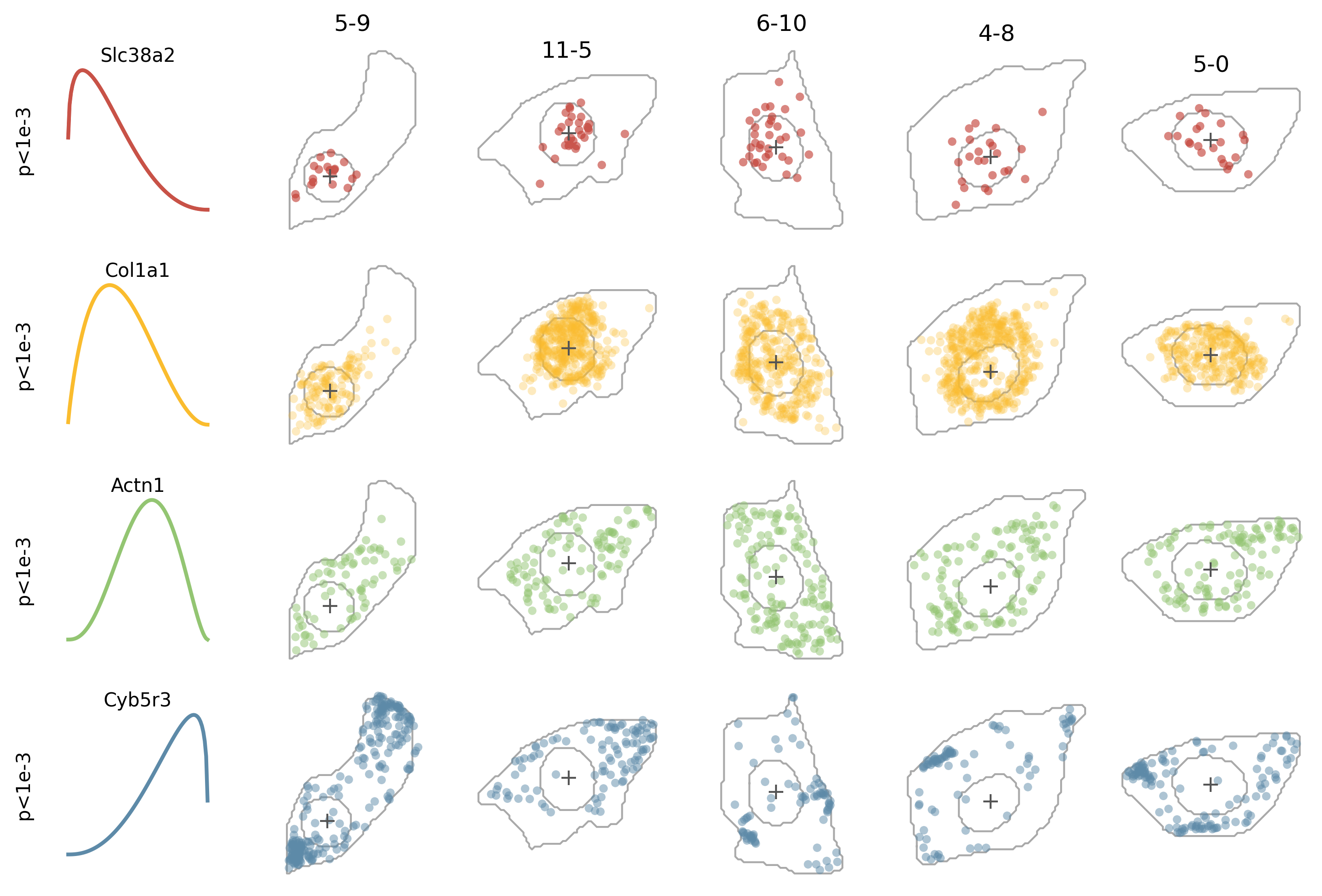
Here Slc38a2 looks like a nuclear localized genes as its estimated expression intensity is high when the relative position is near zero (corresponding to nuclear center); Col1a1 could be a nuclear edge localized gene as its expression intensity peaks around relative position 0.3; Actn1 should be a cytoplasmic localized gene as its expression intensity peak around 0.6; and Cyb5r3 should be a cell membrane localized gene as its expression intensity peaks near 1 (corresponding to the cell membrane).
More to plot: We can further plot the cells and genes to have a more intuitive sense of the localization patterns.
import alphashape
# load intput data
input_data = pd.read_pickle('your_dir/ELLA/scripts/demo/mini_demo/mini_demo_data.pkl')
cell_type = 'fibroblast'
genes = input_data['genes'][cell_type]
cells = input_data['cells'][cell_type]
expr = input_data['expr']
cell_seg = input_data['cell_seg']
nucleus_seg = input_data['nucleus_seg']
# plot
alphas = [0.6, 0.3, 0.5, 0.5]
nr = len(genes)
nc = len(cells)+1
ss_nr = 2
ss_nc = 2
fig = plt.figure(figsize=(nc*ss_nc, nr*ss_nr), dpi=300)
gs = fig.add_gridspec(nr, nc,
width_ratios=[1]*nc,
height_ratios=[1]*nr)
gs.update(wspace=0.0, hspace=0.0)
for j, g in enumerate(genes):
for i, c in enumerate(cells):
if i==0:
ax = plt.subplot(gs[j,i])
lam_est = lam_ests[j]
lam_std = (lam_est-np.min(lam_est))/(np.max(lam_est)-np.min(lam_est)+1e-10)
ax.plot(np.linspace(0,1,len(lam_std)), lam_std, lw=2, color=colors[j])
ax.set_title(g)
#ax.set_ylabel(f'{p_fdr[j]:.3f}')
ax.set_xticks([-0.2,0.5,1.2], [-0.2,0.5,1.2])
ax.set_yticks([-0.2,0.5,1.2], [-0.2,0.5,1.2])
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.spines['bottom'].set_visible(False)
ax.set_xticks([])
ax.set_yticks([])
ax = plt.subplot(gs[j,i+1])
cell_seg_c = cell_seg[cell_seg.cell==c]
nucleus_seg_c = nucleus_seg[nucleus_seg.cell==c]
expr_c = expr[expr.cell==c]
# cell segmentation
x_reduced = (cell_seg_c.x.values//10) * 10 # reduce resolution to speedup alphashape
y_reduced = (cell_seg_c.y.values//10) * 10
points = np.stack((x_reduced, y_reduced)).transpose()
unique_points = np.unique(points, axis=0)
alpha_shape_ = alphashape.alphashape(unique_points, 0.1)
cb_x_, cb_y_ = alpha_shape_.exterior.xy
ax.plot(cb_x_, cb_y_,
alpha=0.5,
color=darkgray, lw=1, zorder=1)
# nuclear segmentation
x_reduced = (nucleus_seg_c.x.values//10) * 10 # reduce res to speedup alphashape
y_reduced = (nucleus_seg_c.y.values//10) * 10
points = np.stack((x_reduced, y_reduced)).transpose()
unique_points = np.unique(points, axis=0)
alpha_shape_ = alphashape.alphashape(unique_points, 0.1)
cb_x_, cb_y_ = alpha_shape_.exterior.xy
ax.plot(cb_x_, cb_y_,
alpha=0.5,
color=darkgray, lw=1, zorder=1)
# gene expr
expr_c_g = expr_c[expr_c.gene==g]
ax.scatter(expr_c_g.x,
expr_c_g.y,
s = 20,
edgecolor='none',
color=colors[j],
alpha=alphas[j],
zorder=2)
# cell center
xc = expr_c.centerX.iloc[0]
yc = expr_c.centerY.iloc[0]
ax.scatter(xc, yc, c=darkgray, marker='+',lw=1, s=60, zorder=3)
ax.set_aspect('equal', adjustable='box')
#ax.axis('off')
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
ax.spines['left'].set_visible(False)
ax.spines['bottom'].set_visible(False)
ax.set_xticks([])
ax.set_yticks([])
if j==0:
ax.set_title(c)