A Complete Demo
Here’s a demo introducing a complete ELLA analysis pipeline.
The script that will be used in this demo should have already been downloaded (while cloning the ELLA repo). The data (complete_demo_data.pkl) that will be used can be downloaded from here. You should be able to orgnize these at your local ELLA folder as follows:
ELLA/scripts/demo/complete_demo/
├── lightning_logs
│ └── run1
├── log
├── complete_demo_data.pkl
├── complete_demo_postprocess.ipynb
├── prepared_data
└── run_complete_demo.sh
The data is a subset of the processed seqFISH+ embryonic fibroblast dataset. The input data (complete_demo_data.pkl) mainly contains a dictionary of three dataframes corresponding to gene expression, cell segmentation, and nucleus segmentation (optional) with 20 cells and 50 genes.
ELLA Anlysis
- Preprocess
python -m ella.data.prepare_data -i your_dir/ELLA/scripts/demo/complete_demo/complete_demo_data.pkl -o your_dir/ELLA/scripts/demo/complete_demo/prepared_data - Run ELLA
bash run_complete_demo.shThe corresponding recipe is
complete_demo.yaml. - Postprocess
Usingcomplete_demo_postprocess.ipynb.
Specifically, we can now cluster the estimated (significant) expression intensities into clusters of patterns. We find the optimal number of kmeans clusters K with the ELBOW method where K is chosen as a point where the distortion/inertia begins to decrease more slowly.
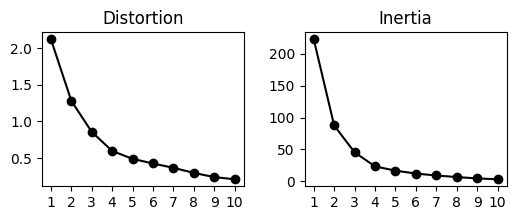
Based on the plots, it seems 5 can be a proper choice, thus let’s proceed with K=5 to obtain 5 pattern clusters:
Pattern 1: 12 genes
Pattern 2: 7 genes
Pattern 3: 11 genes
Pattern 4: 9 genes
Pattern 5: 6 genes
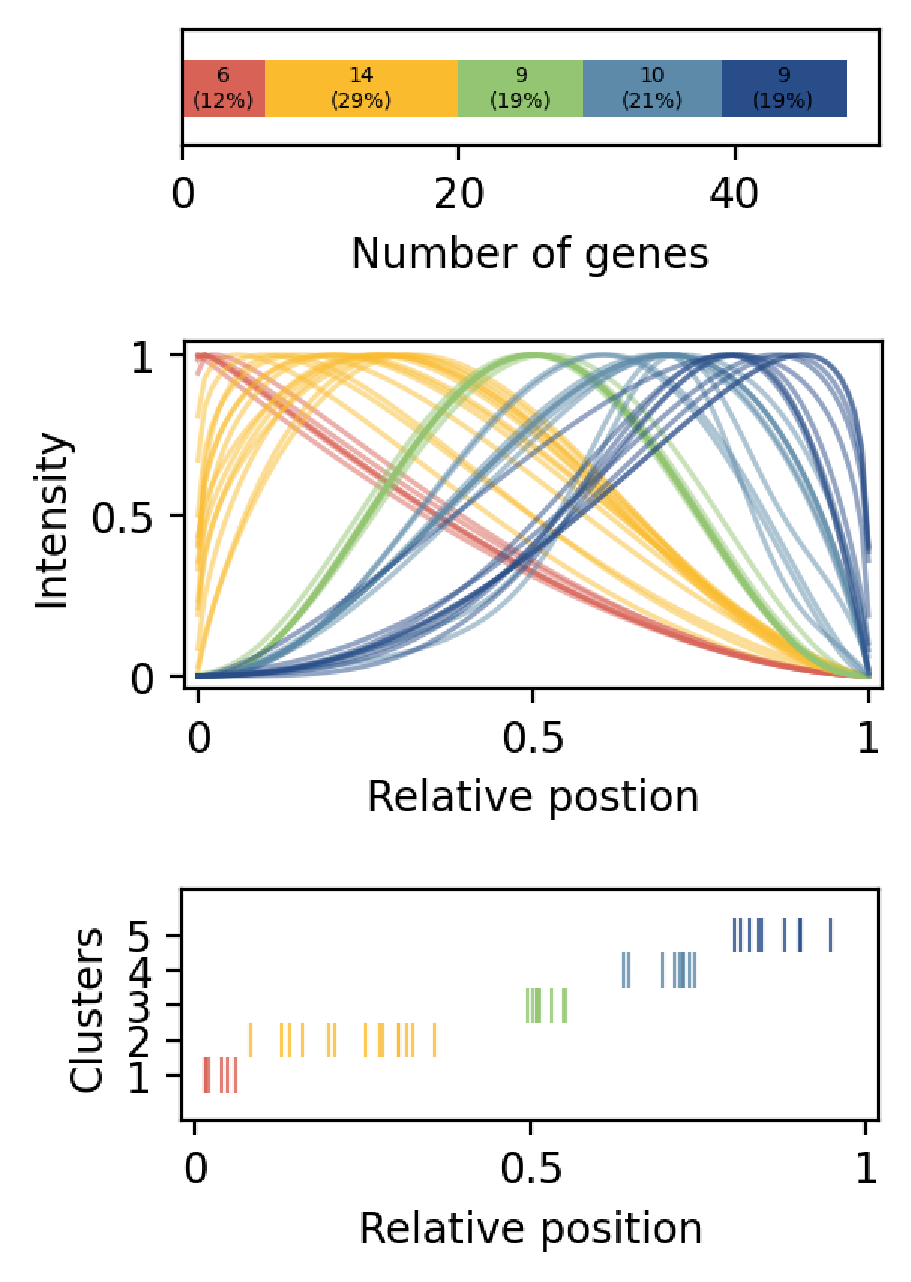
Plots: numbers and proportions of significant genes, estimated expression patterns, and estimated pattern scores
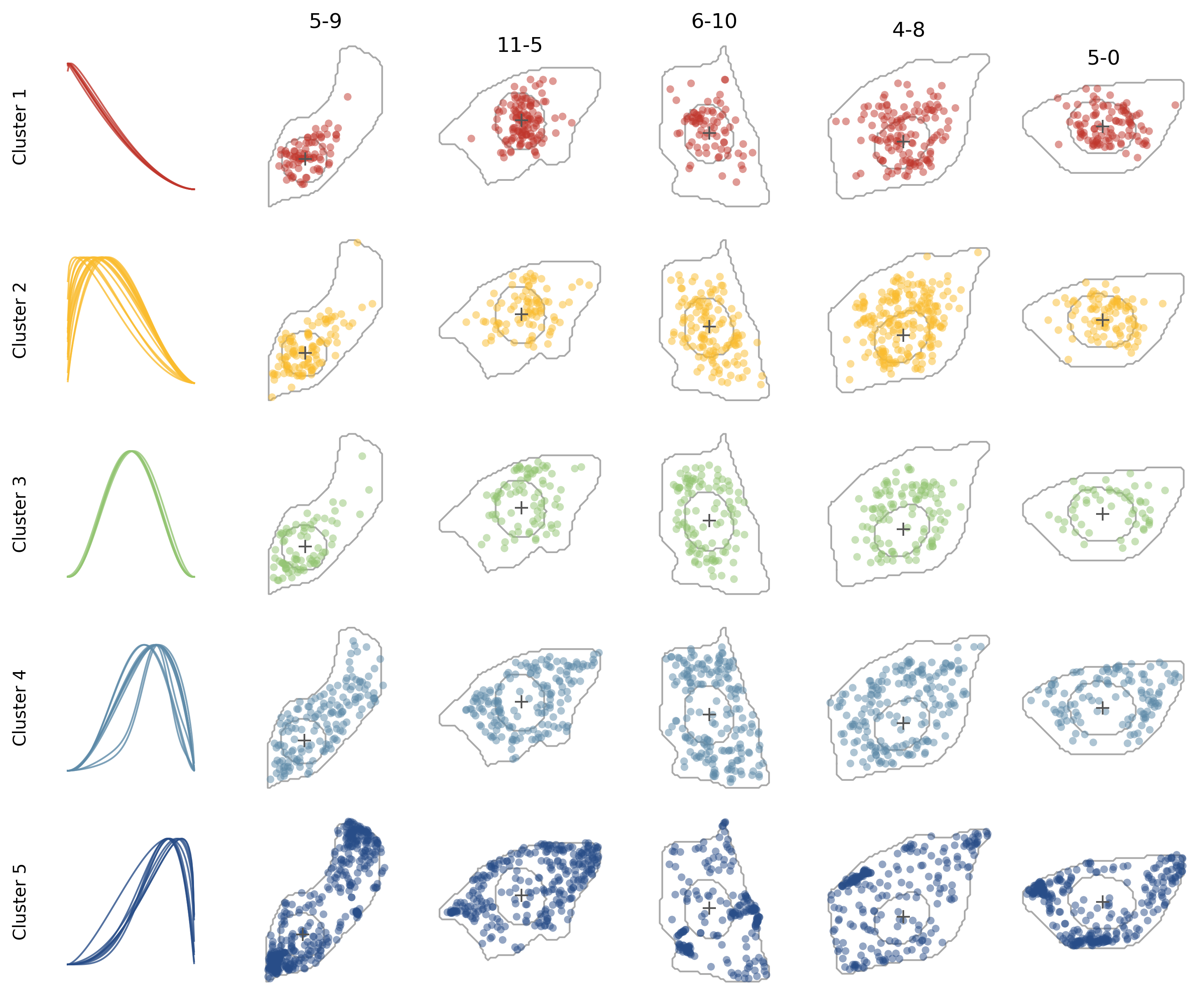
We can overlay all genes in the same cluster in cells to have a more intuitive sense of the patterns.