Welcome to ELLA’s page!
What’s ELLA for?
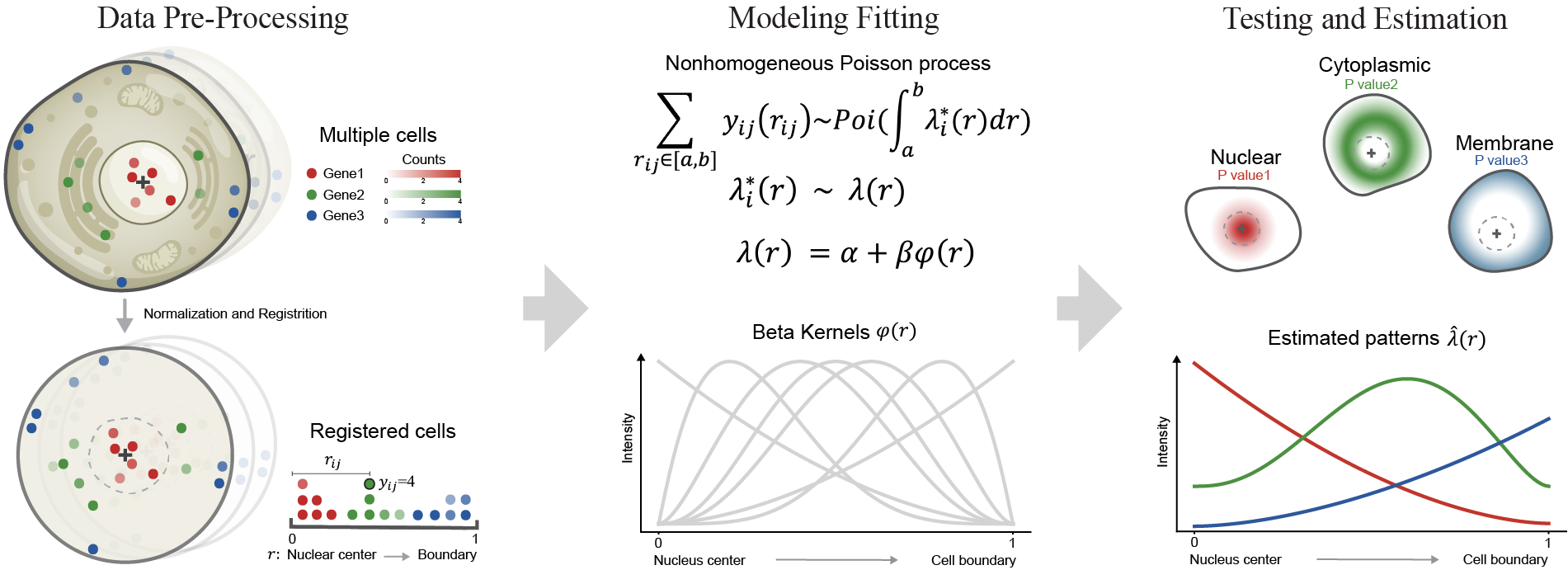
ELLA (subcellular Expression LocaLization Analysis) is a statistical method that integrates high-resolution spatially resolved gene expression data with histology imaging data to identify the subcellular mRNA localization patterns in various spatially resolved transcriptomic techniques.
What’s the model?
ELLA models spatial count data through a (over-dispersed) nonhomogeneous Poisson process model and relies on an expression gradient function to characterize the subcellular mRNA localization pattern, producing effective control of type I errors and yielding high statistical power.
How to try it out?
ELLA is implemented in python with torch. Install ELLA library together with the dependencies follow the steps shown in Install ELLA. After intalling ELLA, this Minimum Demo can be a good start. To take further steps, a complete demo, a showcase of input data, some advanced usages, and the scripts that we used for the real data anlayses in the ELLA manuscript are also shared via this website.
Report issues.
Please let us know if you encounter any issues or have any suggestions or comments. ELLA appreciates your contributions!
Cite ELLA.
Jade Wang, Xiang Zhou. ELLA: Modeling the Subcellular Spatial Variation of Gene Expression within Cells in High-Resolution Spatial Transcriptomics, 2024.
Check out our Zhou Lab website for more softwares :)